Image Hyperparameter Optimization - Jupyter Notebook
hyperparameter_search.ipynb demonstrates using ClearML's HyperParameterOptimizer class to perform automated hyperparameter optimization (HPO).
The code creates a HyperParameterOptimizer object, which is a search controller. The search controller uses the Optuna search strategy optimizer. The example maximizes total accuracy by finding an optimal number of epochs, batch size, base learning rate, and dropout. ClearML automatically logs the optimization's top performing experiments.
The experiment whose hyperparameters are optimized is named image_classification_CIFAR10. It is created by running another
ClearML example, image_classification_CIFAR10.ipynb,
which must run before hyperparameter_search.ipynb.
The optimizer Task, Hyperparameter Optimization, and the experiments appear individually in the ClearML Web UI.
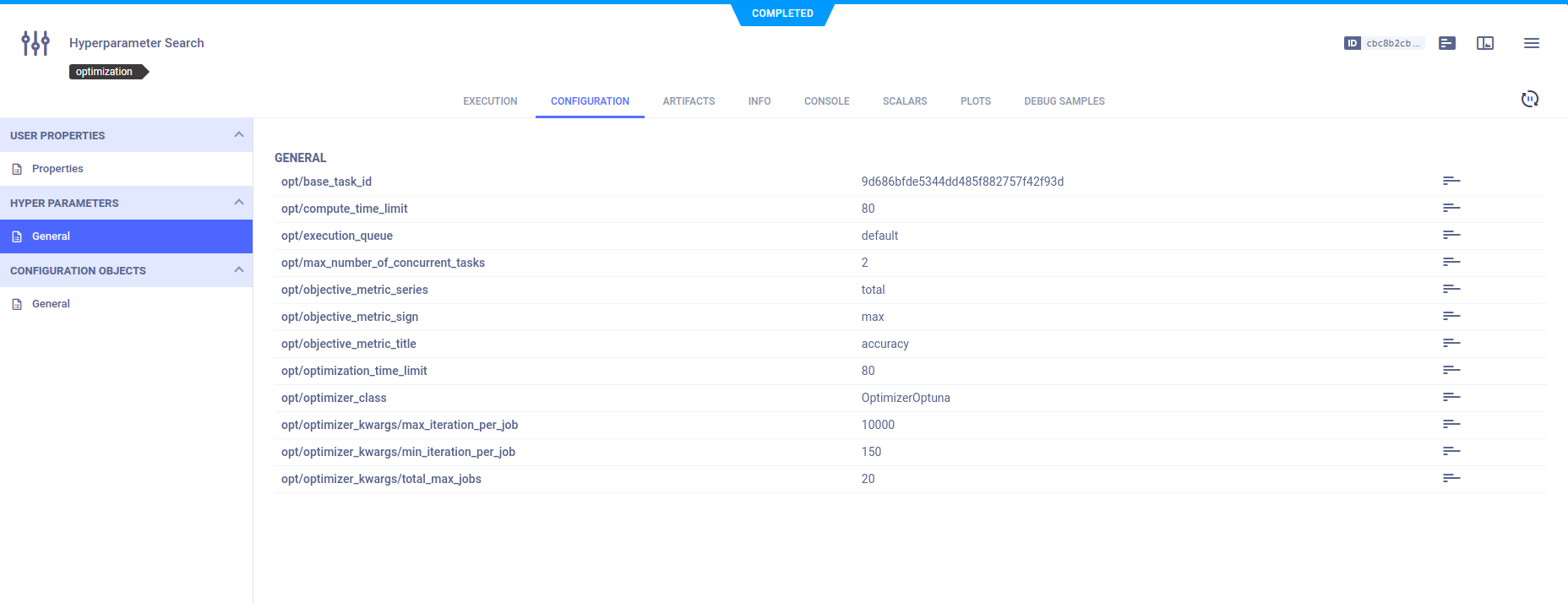
Optimizer Task
Hyperparameters
The HyperParameterOptimizer's configuration, which is provided when the object instantiated, are displayed in the
optimizer task's CONFIGURATION > HYPERPARAMETERS > General section.
optimizer = HyperParameterOptimizer(
base_task_id=TEMPLATE_TASK_ID, # This is the experiment we want to optimize
# here we define the hyperparameters to optimize
hyper_parameters=[
UniformIntegerParameterRange('number_of_epochs', min_value=2, max_value=12, step_size=2),
UniformIntegerParameterRange('batch_size', min_value=2, max_value=16, step_size=2),
UniformParameterRange('dropout', min_value=0, max_value=0.5, step_size=0.05),
UniformParameterRange('base_lr', min_value=0.00025, max_value=0.01, step_size=0.00025),
],
# setting the objective metric we want to maximize/minimize
objective_metric_title='accuracy',
objective_metric_series='total',
objective_metric_sign='max', # maximize or minimize the objective metric
# setting optimizer - clearml supports GridSearch, RandomSearch, OptimizerBOHB and OptimizerOptuna
optimizer_class=OptimizerOptuna,
# Configuring optimization parameters
execution_queue='dan_queue', # queue to schedule the experiments for execution
max_number_of_concurrent_tasks=2, # number of concurrent experiments
optimization_time_limit=60., # set the time limit for the optimization process
compute_time_limit=120, # set the compute time limit (sum of execution time on all machines)
total_max_jobs=20, # set the maximum number of experiments for the optimization.
# Converted to total number of iteration for OptimizerBOHB
min_iteration_per_job=15000, # minimum number of iterations per experiment, till early stopping
max_iteration_per_job=150000, # maximum number of iterations per experiment
)
Console
All console output appears in the optimizer task's CONSOLE.

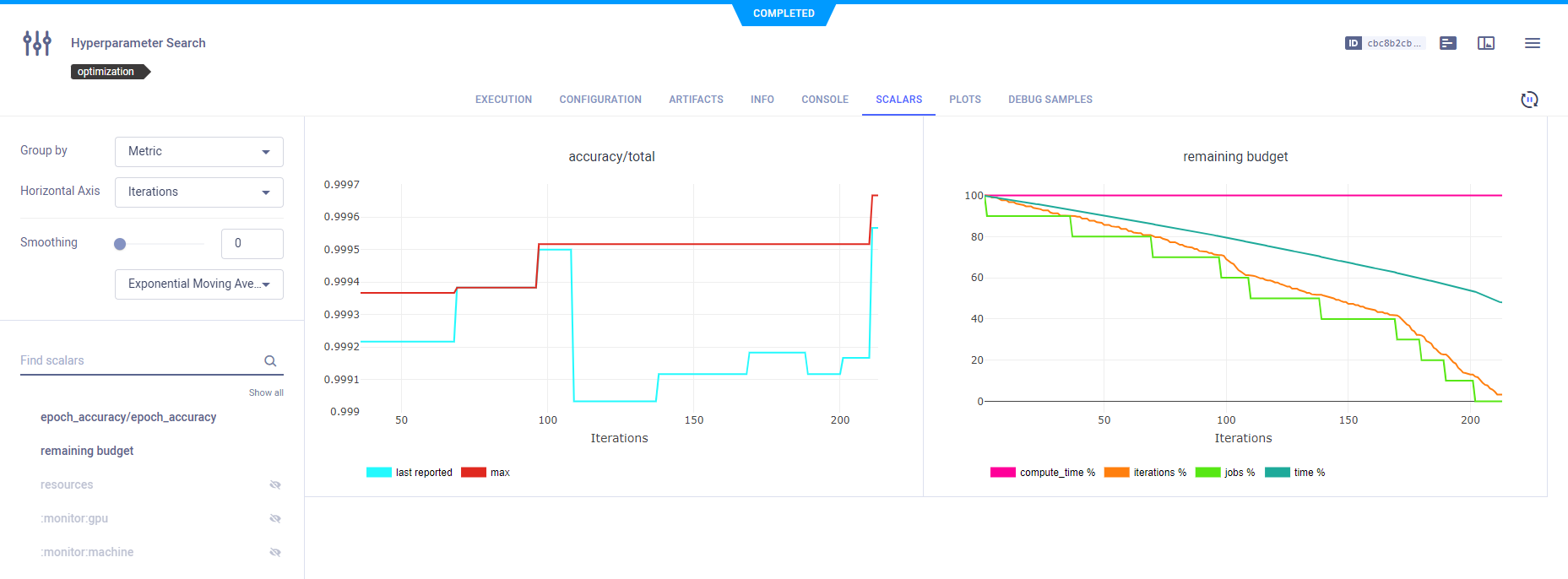
Scalars
Scalar metrics for total accuracy and remaining budget by iteration, and a plot of total accuracy by iteration appear in the experiment's SCALARS tab. Remaining budget indicates the percentage of total iterations for all jobs left before that total is reached.
ClearML automatically reports the scalars generated by HyperParameterOptimizer.
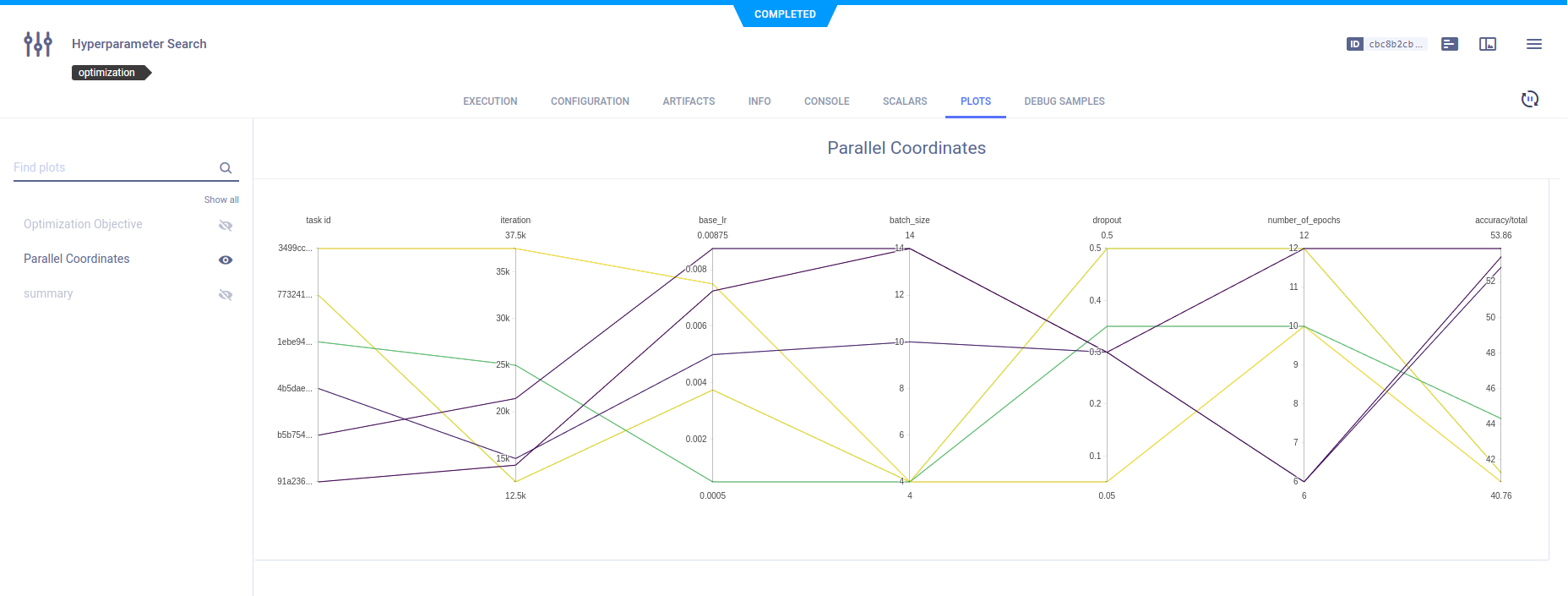
Plots
The optimization task automatically records and monitors the different trial tasks' configuration and execution details, and provides a summary of the optimization results in tabular and parallel coordinate formats. View these plots in the task's PLOTS.


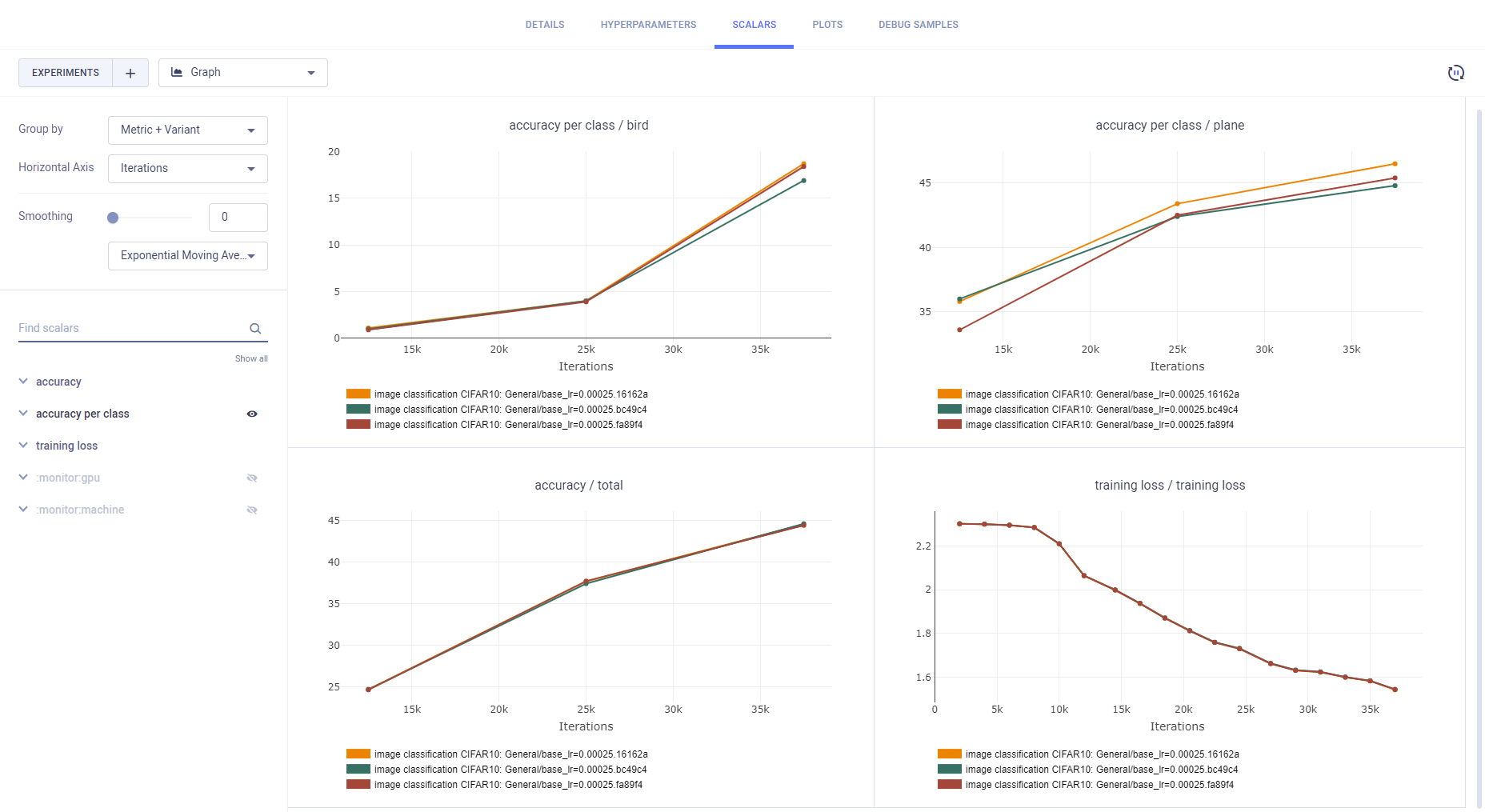
Experiments Comparison
ClearML automatically logs each job, meaning each experiment that executes with a set of hyperparameters, separately. Each appears as an individual experiment in the ClearML Web UI, where the Task name is image_classification_CIFAR10 and the hyperparameters appended.
For example: image_classification_CIFAR10: base_lr=0.0075 batch_size=12 dropout=0.05 number_of_epochs=6.
Compare the experiments' configuration, results, plots, debug samples, and more, using the WebApp comparison features.